Difference between revisions of "BioPortal Import Plugin"
(Creating the page) |
|||
| Line 15: | Line 15: | ||
|Affiliation1=Stanford Center for Biomedical Informatics Research | |Affiliation1=Stanford Center for Biomedical Informatics Research | ||
}} | }} | ||
| + | <div style="display:block; float:left; width:100%;"> | ||
| + | |||
| + | ` | ||
| + | = Overview = | ||
| + | |||
| + | The BioPortal Reference Plugin allows the user '''to insert references to terms, classes, etc. from external ontologies and terminologies''' stored in [http://bioportal.bioontology.org/ BioPortal] into his or her own ontology without the need to import the external ontology. The plugin allows the '''easy navigation''' from the domain ontology in Protege to the external ontology by simply clicking on the generated '''hyperlink of a reference'''. The hyperlink will take the user directly into BioPortal to the referenced term. The user may also search BioPortal for alternative terms. | ||
| + | |||
| + | The plugin is configurable and works with Protege OWL, RDF, Frames, client-server and all backends of Protege. | ||
| + | |||
| + | The plugin is a slot widget that can be attached to any object or annotation property in OWL, or any slot with value type instance, class or any in Frames. | ||
| + | |||
| + | |||
| + | = How does it look like = | ||
| + | |||
| + | Like this: | ||
| + | |||
| + | |||
| + | [[File:BioPortalReferencePluginSmall.png]] | ||
| + | |||
| + | |||
| + | = Under the hood = | ||
| + | |||
| + | To store the internal representation of an external reference, the plugin creates by default an instance of a <code>ExternalReference</code> class. The plugin can be configured to create classes instead of instances for the external references. In OWL, the details of the external references are stored as values of predefined annotation properties (e.g., <code>conceptId</code>, <code>ontologyName</code>, <code>url</code> etc.). | ||
| + | |||
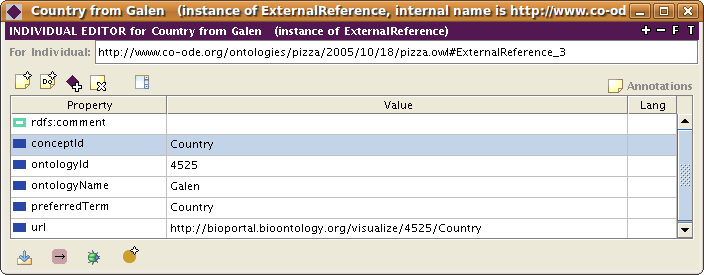
| + | An example of an external reference instance to class <code>Country</code> from the <code>Galen</code> ontology in BioPortal is shown below: | ||
| + | |||
| + | |||
| + | [[File:ExternalReferenceInstance.png]] | ||
| + | |||
| + | |||
| + | To search BioPortal, get the details of a reference, see a graph visualization, etc, the plugin invokes the [http://www.bioontology.org/wiki/index.php/NCBO_REST_services BioPortal Restful Services]. | ||
| + | |||
| + | |||
| + | = How does it work = | ||
| + | |||
| + | The usage of the plugin should be quite straightforward. | ||
| + | |||
| + | '''To create a new external reference''' just click on the link ''Create a new reference from BioPortal''. After that you will see a popup window like this: | ||
| + | |||
| + | |||
| + | [[File:BioPortalSearch.png]] | ||
| + | |||
| + | |||
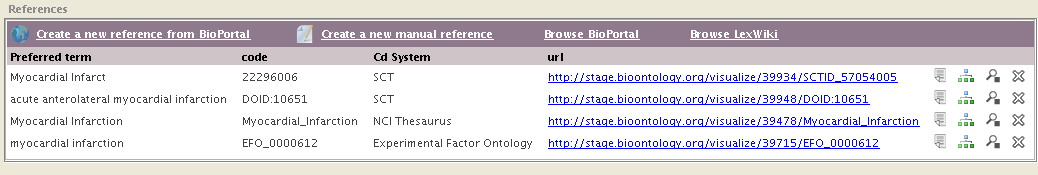
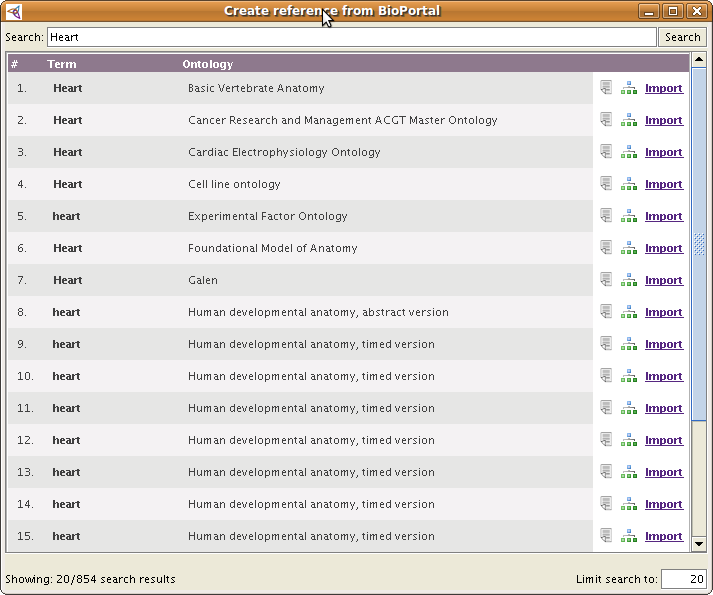
| + | By default, the plugin will start a search in BioPortal using the selected class (or property, individual) name; in the case above, ''Heart''. The panel shows the list of search results for the term from BioPortal: You will see the term name and the ontology in which it was found. | ||
| + | |||
| + | If you are not satisfied with the searched term or results, you may type in your own search term. | ||
| + | |||
| + | '''To import an external reference''' simply click on the ''Import'' link. A new external reference instance (or class, depending on your configuration) has been created for you. | ||
| + | |||
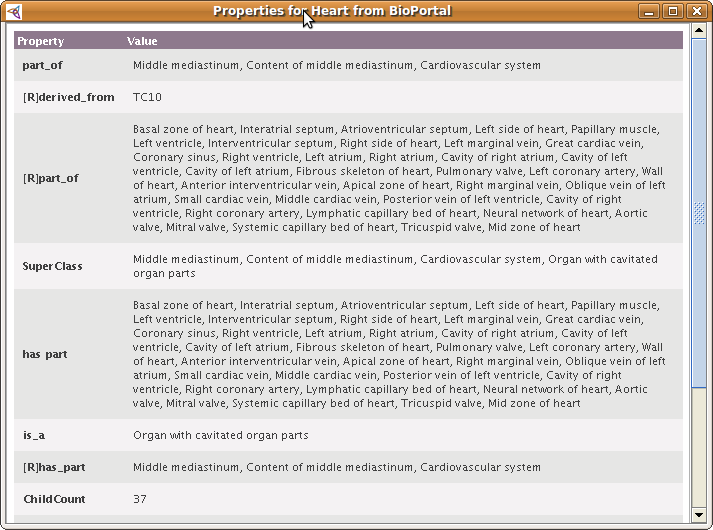
| + | Before importing, you also have the option of '''looking at the content of the term in BioPortal''', by clicking on the little File icon on a row. You will see the details of the term from BioPortal (this will not be imported): | ||
| + | |||
| + | |||
| + | [[File:BioPortalDetails.png]] | ||
| + | |||
| + | |||
| + | Alternatively, you may click on the little tree icon from a row to '''see a graph representation of the neighbourhood of the referenced term''' from BioPortal. A new window will open in your default web browser (e.g. Firefox, Safari, IE, etc.) showing the neighbourhood graph. You will need to have Flash installed to see the graph. | ||
| + | |||
| + | |||
| + | [[File:BioPortalGraph.png]] | ||
| + | |||
| + | |||
| + | = How to use it = | ||
| + | |||
| + | The BioPortalReference plugin is just a slot widget, which means that it can be attached to properties at classes (also metaclasses) to be used instances (or classes). | ||
| + | |||
| + | == Create a reference property == | ||
| + | |||
| + | To use it with OWL projects, first decide on a property that will store as values the instances of external references. You can either choose from existing properties (''foaf:topicOf'', or others), or you can create your own annotation property, for example, ''externalReference''. You may also use object properties, or just plain RDF properties. If you don't already have this property defined in your ontology, please create it. | ||
| + | |||
| + | == Configure the form == | ||
| + | |||
| + | Second step is to configure the form of the class where you want to use the plugin. For example, if you would like to add external references to all classes (e.g., ''Pizza'' class, ''Heart'' class, etc.), then you will have to configure the form of the <code>owl:Class</code>. To configure the form, click on a class in your ontology and click on the little <code>F</code> icon at the top-right hand side of the ClassesTab. You will get a dialog whether to make owl:Class visible, click OK. | ||
| + | |||
| + | Then go to the ''Forms Tab'' (you may already be there after closing the dialog), make sure that owl:Class is selected, and double click on an empty spot. You will get a dialog with all properties and their attached widget. For the reference property from step 1 (e.g., ''externalReference'') associate the ''BioPortalReferenceWidget''. | ||
| + | |||
| + | '''Note''': The new widget will be created under the top widget (it's a bug), so just drag the top widget to right a little bit, and drag from underneath the BioPortalReference widget to a place of your choice in the UI. | ||
| + | |||
| + | |||
| + | == Configure the widget == | ||
| + | |||
| + | Normally, you would not change the default configuration of the widget, but in case you need to customize it for your own project please read this section. Otherwise, you may just skip it. | ||
| + | |||
| + | You may configure the widget in the Forms Tab by double clicking on the BioPortalReference widget. In the '''Columns'' Tab, you may configure what columns and the width of each column that should show up: | ||
| + | |||
| + | |||
| + | [[File:BioPortalColumns.png]] | ||
| + | |||
| + | |||
| + | For example, you may remove the ''url'' slot and this will be removed from the display of the BioPortalReferenceWidget, or you may add other column, or switch the order. | ||
| + | |||
| + | In the '''Search Tab''', you may configure the search parameters for BioPortal: | ||
| + | |||
| + | |||
| + | [[File:BioPortalSearchConfig.png]] | ||
| + | |||
| + | |||
| + | == The result == | ||
| + | |||
| + | Below is a screenshot that shows the OWLClasses tab, for which we have configured using the steps above, the BioPortalReference widget: | ||
| + | |||
| + | |||
| + | [[File:Pizza_with_BPRef.png]] | ||
| + | |||
| + | |||
| + | = Download = | ||
| + | |||
| + | The plugin is distributed with the Protege 3.4.2 release and newer. However, we might need to make changes to the plugin to keep in sync with the BioPortal releases. For this reason, we also provide separate downloads for the plugin. | ||
| + | |||
| + | We have a patch version of the plugin for Protege 3.4.3, that can be downloaded from [http://smi-protege.stanford.edu/collab-protege/tmp/bioportalReference.zip here]. Unzip it into your Protege 3.4.x plugin folder. (Make sure you override the old versions of the plugin.) | ||
| + | |||
| + | The source code of the plugin is available in our SVN: http://smi-protege.stanford.edu/svn/bioportal-reference-plugin/trunk/ and it is licensed under Mozilla Public License. | ||
| + | |||
| + | |||
| + | = Developer's Guide = | ||
| + | |||
| + | This plugin is very easy to extend. | ||
| + | |||
| + | == Configure the classpath and Eclipse setup == | ||
| + | |||
| + | First, you should get the sources of the plugin from the SVN repository, as explained in the above section. The plugin depends on: | ||
| + | * standard-extensions (get the one in the zip from [http://smi-protege.stanford.edu/collab-protege/tmp/bioportalReference.zip here]) | ||
| + | * protege-owl | ||
| + | * xstream libs (get the binary version from [http://xstream.codehaus.org/download.html here], Libs needed in the classpath: xpp3_min_xyz.jar and xstream-xyz.jar) | ||
| + | |||
| + | If you would like to use Eclipse development environment, we have a guide of how to set it up for plugin development [[SetUpEclipseForPlugin|here]]. | ||
| + | |||
| + | == Explanation of classes == | ||
| + | |||
| + | The plugin is very simple. It has classes for UI and classes for accessing BioPortal through rest services. | ||
| + | |||
| + | The slot plugin class (main UI) is: <code>edu.stanford.bmir.protegex.bp.ref.BioPortalReferenceWidget</code> | ||
| + | |||
| + | The panel in which you can search and import from BioPortal is: <code>edu.stanford.bmir.protegex.bp.ref.SearchPanel</code> | ||
| + | |||
| + | The reference model class that creates the reference instances and makes the actual import is: <code>edu.stanford.bmir.protegex.bp.ref.ReferenceModel</code> | ||
| + | |||
| + | If you would like to change the way that references are imported, change the method (or create your own): | ||
| + | |||
| + | public Instance createReference(String bpBaseUrl, String conceptId, String ontologyVersionId, String preferredName, String ontologyName) | ||
| + | |||
| + | The package <code>org.ncbo.stanford</code> contains the classes that can can invoke a rest service to BioPortal and that can read in the results. All you need to invoke the services is in the <code>org.ncbo.stanford.util</code> package. | ||
| + | |||
| + | |||
| + | == Invoke BioPortal Rest services == | ||
| + | |||
| + | The BioPortal Rest services are documented [http://www.bioontology.org/wiki/index.php/NCBO_REST_services here]. | ||
| + | |||
| + | For example, to invoke a search in BioPortal, you can use the utility class: | ||
| + | |||
| + | BioportalSearch sd = new BioportalSearch(); | ||
| + | String urlStr = "http://rest.bioontology.org/bioportal/search/Myocardial%20Infarct?pagesize=20&pagenum=1"; | ||
| + | try { | ||
| + | Page p = sd.getSearchResults(new URL(urlStr)); | ||
| + | SearchResultListBean data = p.getContents(); | ||
| + | for (SearchBean searchBean : data.getSearchResultList()) { | ||
| + | System.out.println(searchBean.getPreferredName() + "\t" + searchBean.getOntologyDisplayLabel()); | ||
| + | } | ||
| + | } catch (MalformedURLException e) { | ||
| + | e.printStackTrace(); | ||
| + | } catch (IOException e) { | ||
| + | e.printStackTrace(); | ||
| + | } | ||
| + | |||
| + | For an example implementation see: <code>edu.stanford.bmir.protegex.bp.ref.SearchPanel.getSearchResultsHtml()</code> | ||
| + | |||
| + | |||
| + | To get all details of a concept, you can use the following code: | ||
| + | |||
| + | BioportalConcept c = new BioportalConcept(); | ||
| + | String urlStr = "http://rest.bioontology.org/bioportal/concepts/39002/BRO:Resource"; | ||
| + | try { | ||
| + | ClassBean cb = c.getConceptProperties(new URL(urlStr)); | ||
| + | System.out.println(cb.getFullId() + " " + cb.getId() + " " + cb.getLabel()); | ||
| + | Map<Object, Object> relationsMap = cb.getRelations(); | ||
| + | for (Map.Entry<Object, Object> e : relationsMap.entrySet()) { | ||
| + | System.out.println(e.getKey() + ": " + e.getValue()); | ||
| + | } | ||
| + | } catch (MalformedURLException e) { | ||
| + | e.printStackTrace(); | ||
| + | } | ||
| + | |||
| + | |||
| + | = Where to get help and complain = | ||
| + | |||
| + | If you have any problems with the plugin, please post a message on the protege mailing list (protege-owl for OWL ontologies, and protege-discussion for Protege frames ontologies). Instructions for posting on the lists are here: | ||
| + | |||
| + | http://protege.stanford.edu/community/lists.html | ||
| + | |||
| + | '''Before posting, please check the email list archives, whether the question has already been answered:''' | ||
| + | |||
| + | http://protege.stanford.edu/community/archives.html | ||
| + | |||
| + | '''Thank you!''' | ||
Revision as of 17:28, April 11, 2011
BioPortal Import Plugin
The BioPortal Import Plugin allows the user to insert into the ontology references to external ontologies and terminologies stored in BioPortal. The user can import an entire tree of classes with a desired depth and choose which properties to import for each class.
Contents
Versions & Compatibility
This section lists available versions of BioPortal Import Plugin.
No version information available.
If you click on the button below to add a new version of BioPortal Import Plugin, you will be asked to define a page title for the new version. Please adhere to the naming convention of BioPortal Import Plugin X.X.X when you define the new page!
Changelog
No version information available.
`
Overview
The BioPortal Reference Plugin allows the user to insert references to terms, classes, etc. from external ontologies and terminologies stored in BioPortal into his or her own ontology without the need to import the external ontology. The plugin allows the easy navigation from the domain ontology in Protege to the external ontology by simply clicking on the generated hyperlink of a reference. The hyperlink will take the user directly into BioPortal to the referenced term. The user may also search BioPortal for alternative terms.
The plugin is configurable and works with Protege OWL, RDF, Frames, client-server and all backends of Protege.
The plugin is a slot widget that can be attached to any object or annotation property in OWL, or any slot with value type instance, class or any in Frames.
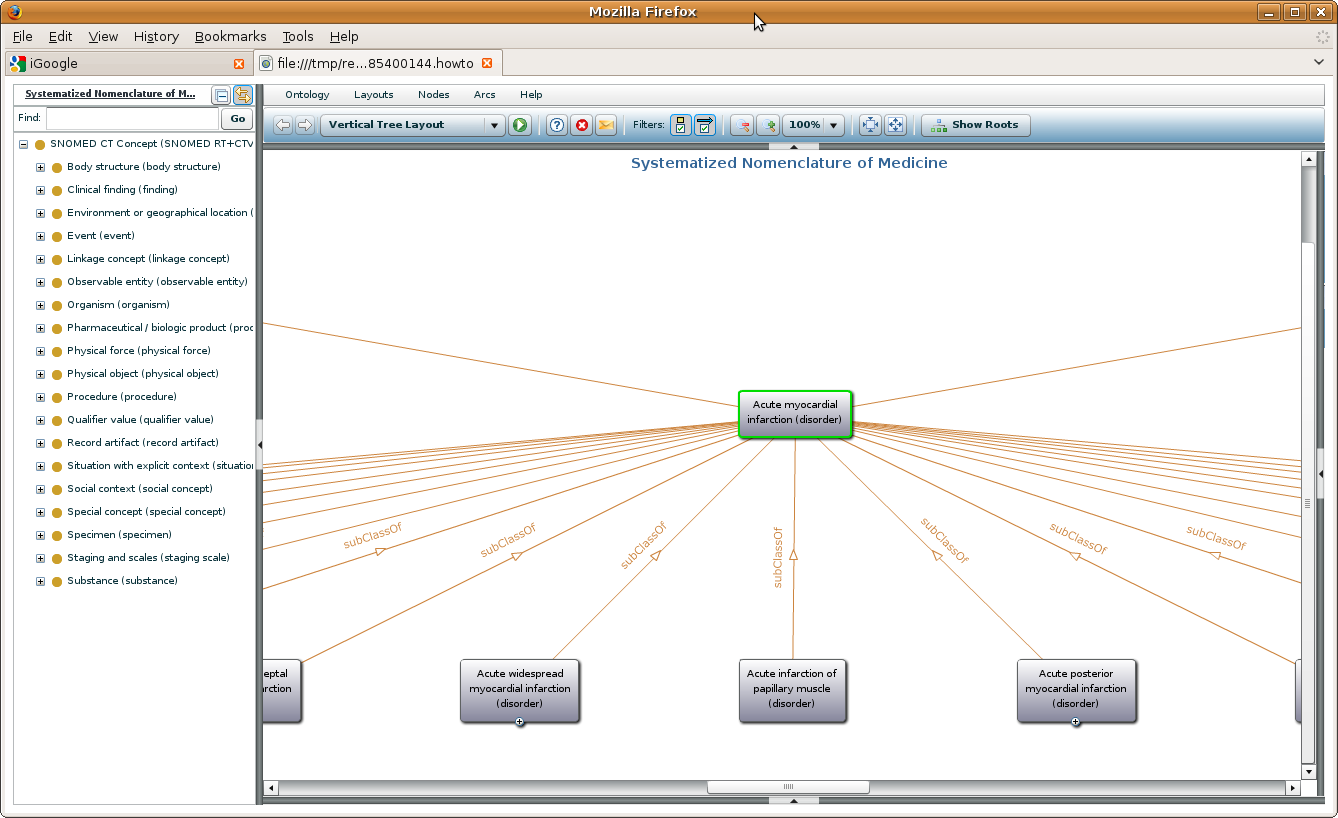
How does it look like
Like this:
Under the hood
To store the internal representation of an external reference, the plugin creates by default an instance of a ExternalReference class. The plugin can be configured to create classes instead of instances for the external references. In OWL, the details of the external references are stored as values of predefined annotation properties (e.g., conceptId, ontologyName, url etc.).
An example of an external reference instance to class Country from the Galen ontology in BioPortal is shown below:
To search BioPortal, get the details of a reference, see a graph visualization, etc, the plugin invokes the BioPortal Restful Services.
How does it work
The usage of the plugin should be quite straightforward.
To create a new external reference just click on the link Create a new reference from BioPortal. After that you will see a popup window like this:
By default, the plugin will start a search in BioPortal using the selected class (or property, individual) name; in the case above, Heart. The panel shows the list of search results for the term from BioPortal: You will see the term name and the ontology in which it was found.
If you are not satisfied with the searched term or results, you may type in your own search term.
To import an external reference simply click on the Import link. A new external reference instance (or class, depending on your configuration) has been created for you.
Before importing, you also have the option of looking at the content of the term in BioPortal, by clicking on the little File icon on a row. You will see the details of the term from BioPortal (this will not be imported):
Alternatively, you may click on the little tree icon from a row to see a graph representation of the neighbourhood of the referenced term from BioPortal. A new window will open in your default web browser (e.g. Firefox, Safari, IE, etc.) showing the neighbourhood graph. You will need to have Flash installed to see the graph.
How to use it
The BioPortalReference plugin is just a slot widget, which means that it can be attached to properties at classes (also metaclasses) to be used instances (or classes).
Create a reference property
To use it with OWL projects, first decide on a property that will store as values the instances of external references. You can either choose from existing properties (foaf:topicOf, or others), or you can create your own annotation property, for example, externalReference. You may also use object properties, or just plain RDF properties. If you don't already have this property defined in your ontology, please create it.
Configure the form
Second step is to configure the form of the class where you want to use the plugin. For example, if you would like to add external references to all classes (e.g., Pizza class, Heart class, etc.), then you will have to configure the form of the owl:Class. To configure the form, click on a class in your ontology and click on the little F icon at the top-right hand side of the ClassesTab. You will get a dialog whether to make owl:Class visible, click OK.
Then go to the Forms Tab (you may already be there after closing the dialog), make sure that owl:Class is selected, and double click on an empty spot. You will get a dialog with all properties and their attached widget. For the reference property from step 1 (e.g., externalReference) associate the BioPortalReferenceWidget.
Note: The new widget will be created under the top widget (it's a bug), so just drag the top widget to right a little bit, and drag from underneath the BioPortalReference widget to a place of your choice in the UI.
Configure the widget
Normally, you would not change the default configuration of the widget, but in case you need to customize it for your own project please read this section. Otherwise, you may just skip it.
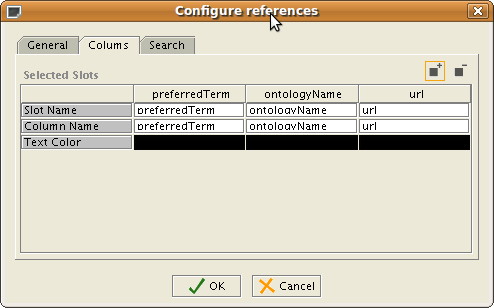
You may configure the widget in the Forms Tab by double clicking on the BioPortalReference widget. In the 'Columns Tab, you may configure what columns and the width of each column that should show up:
For example, you may remove the url slot and this will be removed from the display of the BioPortalReferenceWidget, or you may add other column, or switch the order.
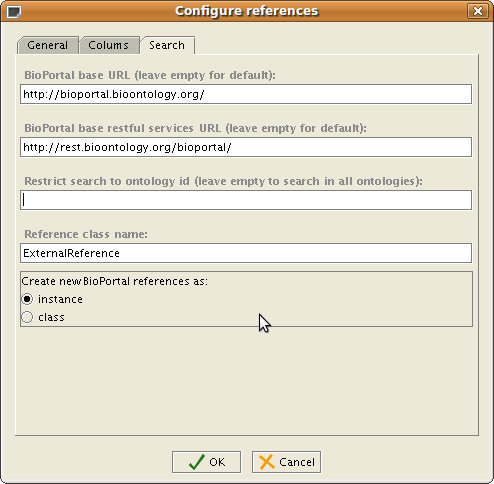
In the Search Tab, you may configure the search parameters for BioPortal:
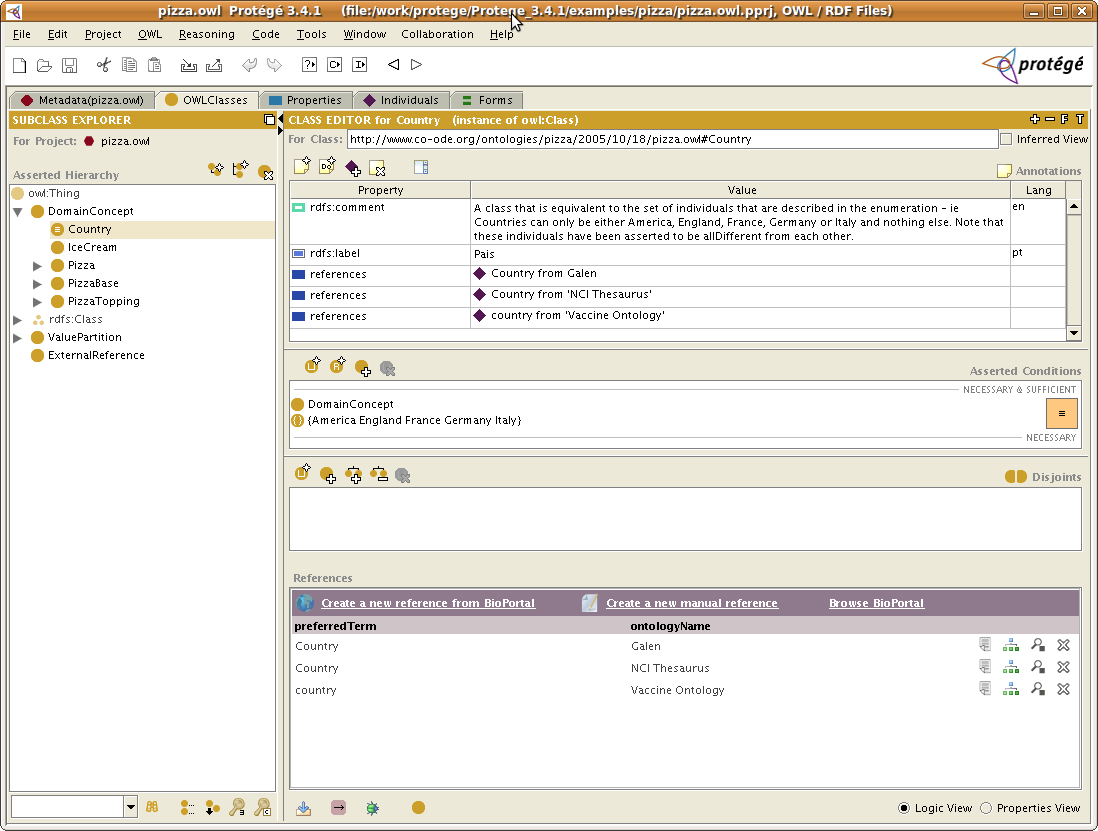
The result
Below is a screenshot that shows the OWLClasses tab, for which we have configured using the steps above, the BioPortalReference widget:
Download
The plugin is distributed with the Protege 3.4.2 release and newer. However, we might need to make changes to the plugin to keep in sync with the BioPortal releases. For this reason, we also provide separate downloads for the plugin.
We have a patch version of the plugin for Protege 3.4.3, that can be downloaded from here. Unzip it into your Protege 3.4.x plugin folder. (Make sure you override the old versions of the plugin.)
The source code of the plugin is available in our SVN: http://smi-protege.stanford.edu/svn/bioportal-reference-plugin/trunk/ and it is licensed under Mozilla Public License.
Developer's Guide
This plugin is very easy to extend.
Configure the classpath and Eclipse setup
First, you should get the sources of the plugin from the SVN repository, as explained in the above section. The plugin depends on:
- standard-extensions (get the one in the zip from here)
- protege-owl
- xstream libs (get the binary version from here, Libs needed in the classpath: xpp3_min_xyz.jar and xstream-xyz.jar)
If you would like to use Eclipse development environment, we have a guide of how to set it up for plugin development here.
Explanation of classes
The plugin is very simple. It has classes for UI and classes for accessing BioPortal through rest services.
The slot plugin class (main UI) is: edu.stanford.bmir.protegex.bp.ref.BioPortalReferenceWidget
The panel in which you can search and import from BioPortal is: edu.stanford.bmir.protegex.bp.ref.SearchPanel
The reference model class that creates the reference instances and makes the actual import is: edu.stanford.bmir.protegex.bp.ref.ReferenceModel
If you would like to change the way that references are imported, change the method (or create your own):
public Instance createReference(String bpBaseUrl, String conceptId, String ontologyVersionId, String preferredName, String ontologyName)
The package org.ncbo.stanford contains the classes that can can invoke a rest service to BioPortal and that can read in the results. All you need to invoke the services is in the org.ncbo.stanford.util package.
Invoke BioPortal Rest services
The BioPortal Rest services are documented here.
For example, to invoke a search in BioPortal, you can use the utility class:
BioportalSearch sd = new BioportalSearch(); String urlStr = "http://rest.bioontology.org/bioportal/search/Myocardial%20Infarct?pagesize=20&pagenum=1"; try { Page p = sd.getSearchResults(new URL(urlStr)); SearchResultListBean data = p.getContents(); for (SearchBean searchBean : data.getSearchResultList()) { System.out.println(searchBean.getPreferredName() + "\t" + searchBean.getOntologyDisplayLabel()); } } catch (MalformedURLException e) { e.printStackTrace(); } catch (IOException e) { e.printStackTrace(); }
For an example implementation see: edu.stanford.bmir.protegex.bp.ref.SearchPanel.getSearchResultsHtml()
To get all details of a concept, you can use the following code:
BioportalConcept c = new BioportalConcept(); String urlStr = "http://rest.bioontology.org/bioportal/concepts/39002/BRO:Resource"; try { ClassBean cb = c.getConceptProperties(new URL(urlStr)); System.out.println(cb.getFullId() + " " + cb.getId() + " " + cb.getLabel()); Map<Object, Object> relationsMap = cb.getRelations(); for (Map.Entry<Object, Object> e : relationsMap.entrySet()) { System.out.println(e.getKey() + ": " + e.getValue()); } } catch (MalformedURLException e) { e.printStackTrace(); }
Where to get help and complain
If you have any problems with the plugin, please post a message on the protege mailing list (protege-owl for OWL ontologies, and protege-discussion for Protege frames ontologies). Instructions for posting on the lists are here:
http://protege.stanford.edu/community/lists.html
Before posting, please check the email list archives, whether the question has already been answered:
Thank you!