Difference between revisions of "BioPortal Import Plugin"
m |
|||
| Line 32: | Line 32: | ||
The BioPortal Import plugin is added as a button to the OWL Classes tab of the Protege-OWL 3.x user interface. The user can browse the class tree of the selected BioPortal ontology right from the Protege desktop interface, and also view the properties of each class. | The BioPortal Import plugin is added as a button to the OWL Classes tab of the Protege-OWL 3.x user interface. The user can browse the class tree of the selected BioPortal ontology right from the Protege desktop interface, and also view the properties of each class. | ||
| − | |||
| − | The '''import configuration''' allows the user to customize the import and it includes: | + | The plugin is configurable and works with Protege-OWL. The '''import configuration''' allows the user to customize the import and it includes: |
* import entire subtrees of classes from BioPortal ontologies | * import entire subtrees of classes from BioPortal ontologies | ||
| Line 40: | Line 39: | ||
* choose which (if any) property values to import for each class and assign an equivalent user property for each imported property, if desired | * choose which (if any) property values to import for each class and assign an equivalent user property for each imported property, if desired | ||
* import metadata at the level of a class or the ontology (e.g. import author, timestamp, source ontology and version, etc.) | * import metadata at the level of a class or the ontology (e.g. import author, timestamp, source ontology and version, etc.) | ||
| + | |||
| + | |||
| + | A related plugin to this one that allows users to create reference to classes in BioPortal (rather than importing them) is the '''[[BioPortal_Reference_Plugin|BioPortal Reference Plugin]]'''. | ||
Revision as of 15:18, July 15, 2011
BioPortal Import Plugin
The BioPortal Import Plugin allows the user to import classes from external ontologies stored in BioPortal. The user can import entire trees of classes with a desired depth and choose which properties to import for each class.
Contents
Versions & Compatibility
This section lists available versions of BioPortal Import Plugin.
No version information available.
If you click on the button below to add a new version of BioPortal Import Plugin, you will be asked to define a page title for the new version. Please adhere to the naming convention of BioPortal Import Plugin X.X.X when you define the new page!
Changelog
No version information available.
`
Overview
The plugin allows a user to import classes from external ontologies and terminologies stored in the BioPortal ontology repository into the local ontology.
The basic steps in using the plugin are to:
- browse through the BioPortal ontologies (except remote ones) and select which one to import classes from
- select one or more classes to import from the BioPortal ontology,
- configure the import,
- import them as subclasses of the selected class in the local ontology.
The plugin uses the BioPortal REST services to access and import the content of BioPortal ontologies. However, ontologies denoted as remote in BioPortal cannot be explored in the plugin, since BioPortal does not provide the information for these ontologies through its REST services.
The BioPortal Import plugin is added as a button to the OWL Classes tab of the Protege-OWL 3.x user interface. The user can browse the class tree of the selected BioPortal ontology right from the Protege desktop interface, and also view the properties of each class.
The plugin is configurable and works with Protege-OWL. The import configuration allows the user to customize the import and it includes:
- import entire subtrees of classes from BioPortal ontologies
- save the imported classes as a separate ontology and/or as a file to the local disk
- choose which (if any) property values to import for each class and assign an equivalent user property for each imported property, if desired
- import metadata at the level of a class or the ontology (e.g. import author, timestamp, source ontology and version, etc.)
A related plugin to this one that allows users to create reference to classes in BioPortal (rather than importing them) is the BioPortal Reference Plugin.
What does it look like
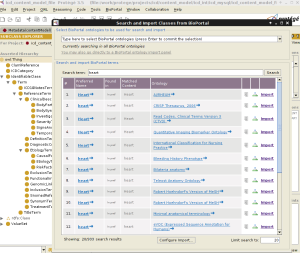
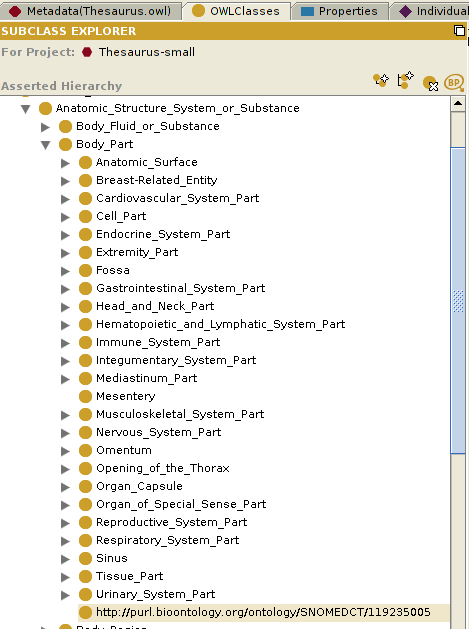
This is a screenshot of the BioPortal Import plugin showing the SNOMED CT ontology classes:
How to use it
The BioPortal Import plugin can be invoked by selecting a class of a domain ontology in the OwlClasses tab in Protege-OWL and then clicking on the icon as shown below:
Importing classes
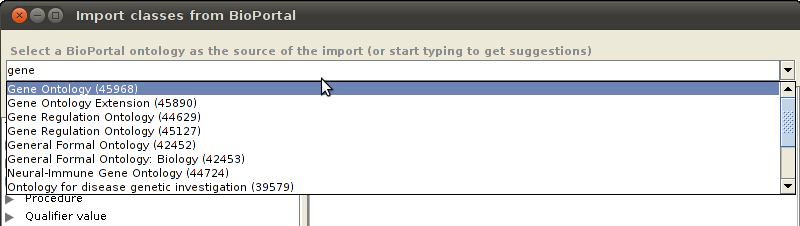
In the plugin window, select a BioPortal ontology from the dropdown list.
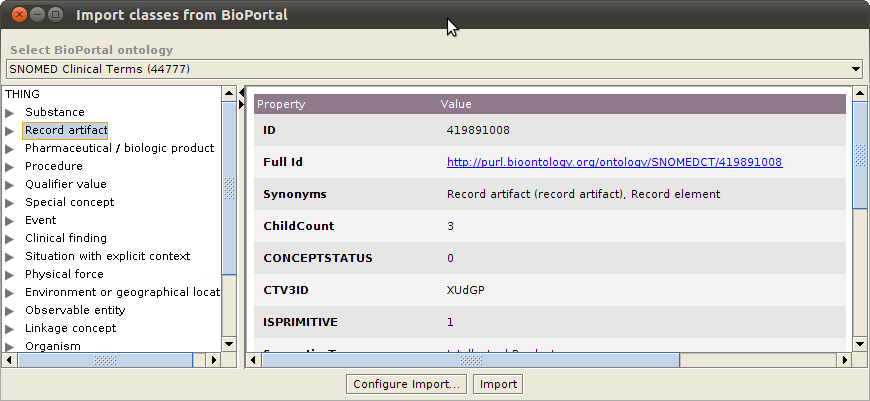
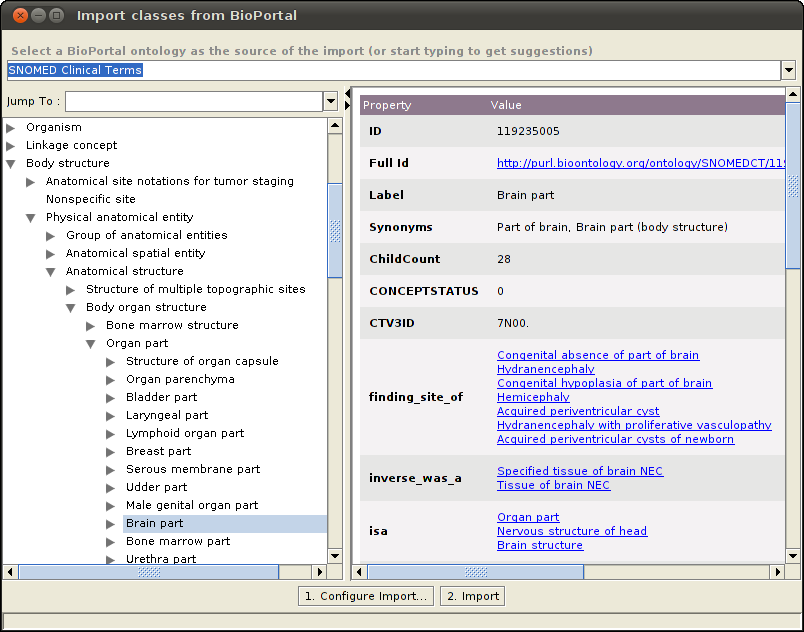
The plugin will show all the classes belonging to the selected ontology in an expandable tree format in the left pane. Selecting one of the classes will show the properties for that class in the details pane. Select one or more classes to import from the BioPortal ontology.
Configure the settings for importing classes by clicking on the "Configure Import..." button. More details about configuring the settings are provided in the "Plugin Configuration" section below.
Click on the "Import" button to import all selected classes into the local ontology.
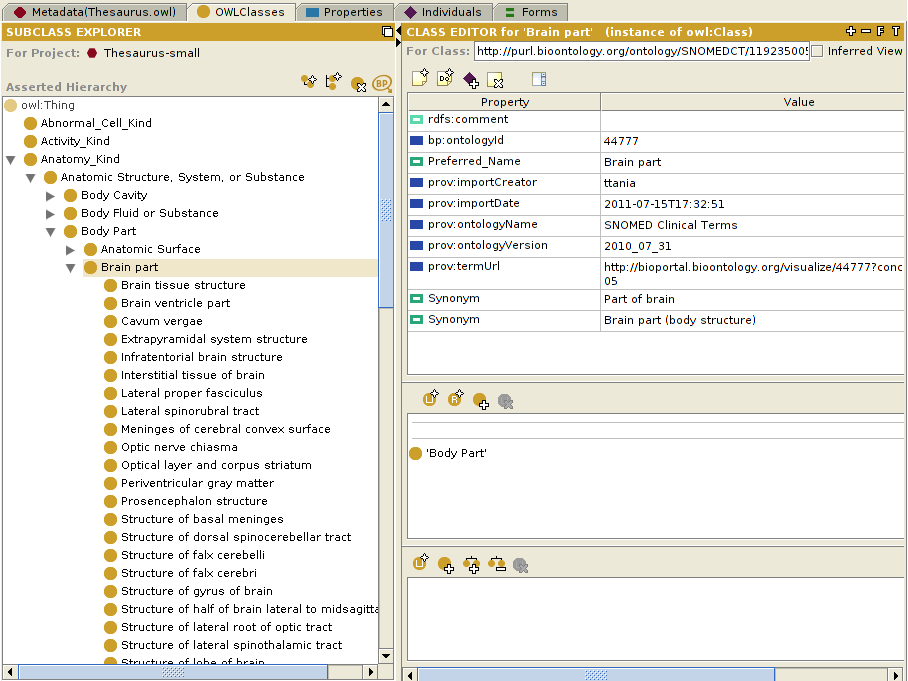
Below is an example of the Mosquito-borne flavivirus hemorrhagic fever class (Term ID 240489005) of the SNOMED CT ontology imported into the Abnormal_Cell class of the NCI Thesaurus ontology.
Plugin configuration
The user can configure the settings for all the above features of the plugin in two ways: 1. selecting "BioPortal>Configure..." from the Protege window menu bar, or 2. clicking on "Configure Import..." in the plugin window. It also allows the user to configure the base URL for BioPortal and BioPortal REST services, if required.
Configuring Imports
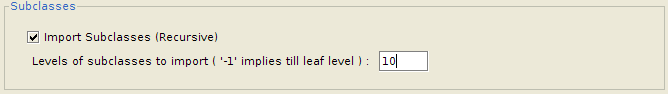
Importing a subtree of classes
If the user wants to import an entire subtree of classes, the plugin provides an option to enable it, as well as to configure the number of levels down the tree to import, starting from the children of the selected class. This makes it very convenient to import entire parts of an ontology at once.
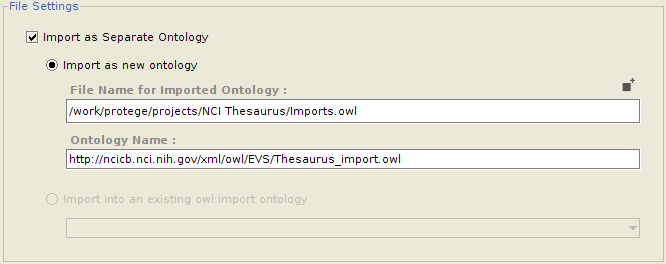
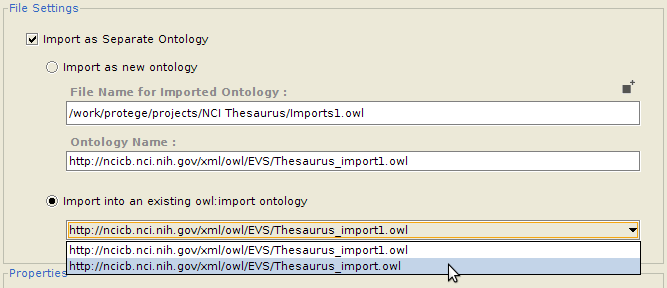
Importing into a separate ontology
Often, a user might want to import classes into a separate ontology, which can then be imported into the main ontology. One advantage of this is that the imported classes can be edited and maintained separately. It also makes removing all the imports altogether much easier. If so desired, the user can configure whether he/she wants to import the selected classes into an entirely new ontology (in which case the ontology name and the name of the file to save it to needs to be provided), or as part of an already imported ontology (for which he needs to select from a list of already imported ontologies which are writable).
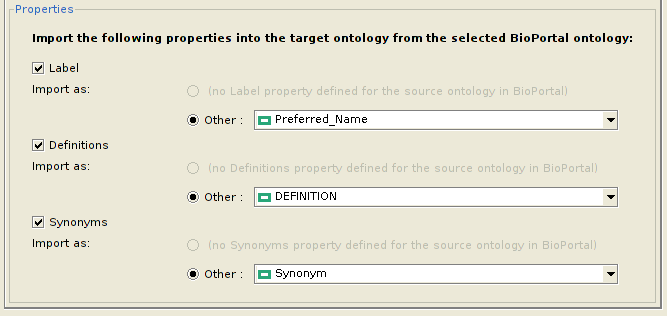
Configuring properties to be imported
Apart from the classes, the user might also wish to import various properties of each class, along with their values. However, since the same property might not exist in the local ontology, the user can choose which property of the local ontology to assign it to. Currently, the plugin allows a user to import only the Label, Definitions and Synonyms properties from BioPortal. However, this list might be extended in the future. The figure below depicts how to configure which properties to import for each class, and their equivalent properties in the local ontology.
The result
Below is a screenshot that shows the NCI Thesaurus ontology from the OWLClasses tab, into which we have imported classes from the SNOMED CT ontology.
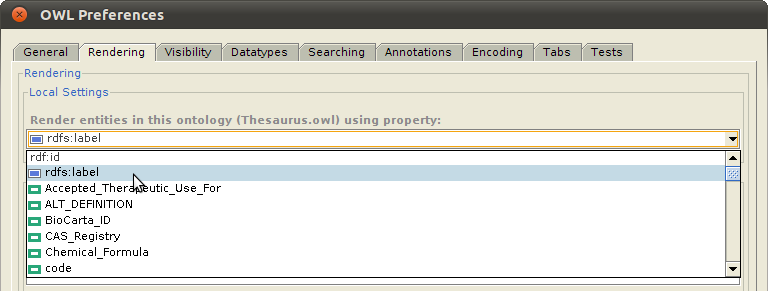
Note: Notice that the highlighted class in the above example, Physical Force (Term ID 78621006) of the SNOMED CT ontology, is one of the classes imported into the NCI Thesaurus ontology, and is displayed using the value of the label property, imported as rdfs:label. This is much more intuitive than displaying the default rdf:id property, which has the value http://purl.bioontology.org/ontology/SNOMEDCT/78621006. To enable this, select "OWL>Preferences" from the Protege window menu bar, click on the "Rendering" tab, and set the value of the "Render entities in this ontology using property:" to whichever property that you want to use to display the class.
Under the hood
The plugin creates instances of the OWLNamedClass class to store all the imported classes from BioPortal. If the user chooses to import any property of the imported classes, it is assigned to one of the OWLAnnotationProperties of the current OWLModel, as configured by the user.
To display the list of ontologies in BioPortal, get the details of the classes and their properties etc., the plugin invokes the BioPortal Restful Services.
Download
The plugin is distributed with the Protege 3.4.7 release and newer. However, we might need to make changes to the plugin to keep in sync with the BioPortal releases and to add enhancements and bug fixes. For this reason, we also provide separate download for the plugin.
We have a patch version of the plugin that can be downloaded from here. Unzip it into your Protege 3.4.x plugin folder. (Make sure you override the old versions of the plugin.)
The source code of the plugin is available in our SVN: http://smi-protege.stanford.edu/svn/bioportal-reference-plugin/trunk/ and it is licensed under Mozilla Public License.
Where to get help and complain
If you have any problems with the plugin, please post a message on the protege mailing list (protege-owl for OWL ontologies, and protege-discussion for Protege frames ontologies). Instructions for posting on the lists are here:
http://protege.stanford.edu/community/lists.html
Before posting, please check the email list archives, whether the question has already been answered:
Thank you!